Unravelling chromatin regulation in fly embryos
Max Planck researcher reveal an unexpected functionality of key molecules in embryonic development
Histone acetyltransferases are considered key molecules for the activation of gene transcription. For a long time, their contribution to transcription activation has been associated with their enzymatic function. The laboratory of Nicola Iovino at the Max Planck Institute of Immunobiology and Epigenetics in Freiburg has investigated the role of the two major histone acetyltransferases, CBP and Gcn5, in the activation of the fruit fly's embryonic genome. The team found that these two molecules control the activation of the embryonic genome by regulating different groups of genes. Surprisingly, it was also revealed that the enzymatic activity of CBP and Gcn5 is not necessary for gene activation during this developmental window.
At the beginning of embryonic development, the genome is still silent. This is because the embryo has yet to activate its own genes and relies on proteins and gene products stored by the mother in the first few hours or even days. Once this maternal contribution drops, the zygotic genome activation (ZGA) leads the embryo to a state where it simultaneously activates thousands of its genes, advancing development further.
For this transition into independent life to succeed, numerous processes must be precisely orchestrated within a short time window in the embryo. Nicola Iovino's laboratory at the Max Planck Institute in Freiburg researches how the genome is prepared for this transition and which molecules are involved.
“The DNA is not just naked in the cell nucleus, but coiled around histone protein. These proteins package the DNA. The complex of histones and DNA is called chromatin,” says Nicola Iovino, group leader at the Max Planck Institute in Freiburg. Various chromatin factors can remove or attach small molecules called acetyl groups to the histones. This can compress or loosen the packaging of DNA and, thus, silence or make genes readable. “We want to understand what needs to happen in the early embryo's genome and which factors are necessary for the embryo to activate it,” explains Nicola Iovino.
Histone acetyltransferases regulate different sets of genes
The laboratory uses the fruit fly Drosophila melanogaster, which is particularly well suited for studying epigenetic processes due to its short generation time and ease of genetic manipulation. Using a genetic screening approach in this study, Nicola Iovino's team confirmed the contribution of well-known chromatin factors as Polycomb-group proteins and histone variants for the correct activation of the zygotic genome. Given the laboratory’s expertise in studying chromatin functions in other developmental contexts, the team decided to focus on the role of two important chromatin factors: CBP and Gcn5. By attaching acetyl groups (acetylation), these two histone acetyltransferases (HATs) ensure that the protein beads are less tightly bound to the DNA and that genes can be more easily activated.
"We discovered that CBP and Gcn5 display different functions regulating the embryonic genome. CBP primarily activates 'development genes,' which are responsible for early developmental functions such as pattern formation or cell fate. In contrast, Gcn5 activates more 'housekeeping genes,' which are necessary for maintaining every cell's basic functions. Additionally, they use different lysine substrates, meaning they use different sites of the histone to attach the acetyl group," explains Filippo Ciabrelli, first-author of the study.
Due to this specialisation of CBP and Gcn5 in gene regulation in the young genome, the team expected that these two histone acetyltransferases would primarily accumulate on their respective target genes. However, surprisingly, the acetylation marks of CBP and Gcn5 were found on every gene of the fruit fly embryo during ZGA. “We were puzzled by these results, and we started to reconsider the role of CBP and Gcn5 enzymatic activities at this developmental stage,” says Nicola Iovino.
Enzymatic activity is not necessary for gene activation
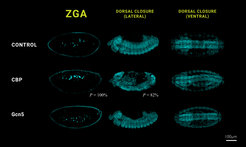
When the team eliminated CBP and Gcn5 or their functional proteins completely through RNA interference, as expected, this led to a failure to initiate ZGA and disrupted embryogenesis. However, when the team generated unique fruit fly embryos that possess catalytically inactive versions of the two histone acetyltransferases, they observed that the embryos were still viable and successfully completed their life cycle. "It even turned out that despite the lack of catalytic activity of CBP and Gcn5, meaning the ability to alter the packaging state of the genetic material in the fly mutants at this developmental stage, the expression of the CBP and Gcn5 target genes was not affected,” explains Nicola Iovino.
To expand the relevance of their observations, the authors collaborated with Vincent Loubiere at the Institute of Molecular Pathology in Vienna. Loubiere’s experimental contribution elucidated how both CBP and Gcn5 proteins can transactivate a reporter gene in-vitro without needing their catalytic activities. “These results confirm what Iovino and colleagues had already observed in their in-vivo model,” says Vincent Loubiere.
Thus, the study demonstrates for the first time that the proteins Gcn5 and CBP play a crucial role in the early development of the fruit fly – even without their catalytic activity. Furthermore, the results show that the enzymatic activity of these key molecules is not necessary for a critical stage of embryonic development and complement the growing body of evidence that some enzymes involved in modifying DNA packaging can function without their catalytic activity.
MR/NI












